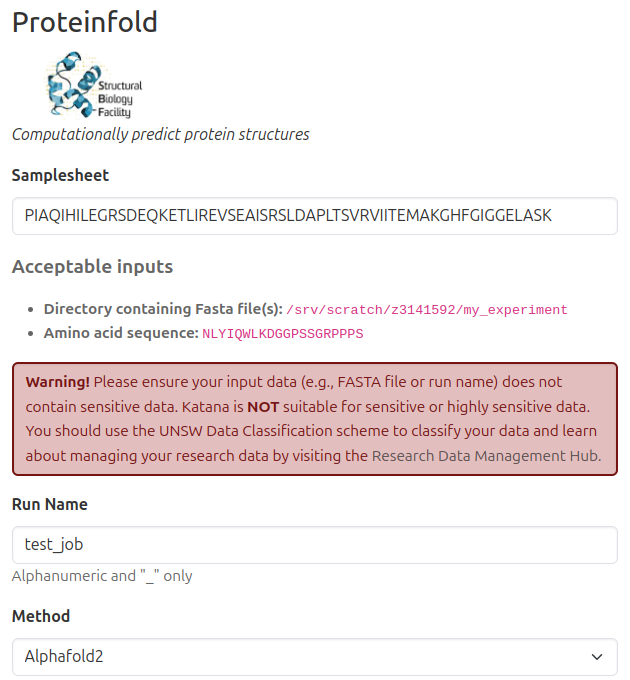
Details
- Numerous structure prediction methods are available via the command line.
- Web services with graphical interfaces are freely available with limited capacity.
- OpenOnDemand provides a simple graphical interface to HPC systems.
- Several institutional HPCs have an OpenOnDemand deployment (e.g. KOD @ UNSW, ARE @ NCI).
Completed
- Pilot app developed and hosted on Katana at UNSW.
In Progress
- Adapt pilot ProteinFold app for ARE at NCI.
Future
- Add support for more optional settings in web form interface.